The distribution of ellagitannins in the plant kingdom is known only for selected plant species and their specific linkage with the plant phylogeny has many gaps in knowledge that need efficient tools to be filled
Large fractionation experiment revealed the main ellagitannins
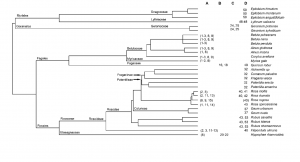
Figure 1. Moilanen et al. (2015) combined the chemotaxonomy of Finnish ellagitannin-producing species with their phylogeny. Especially the occurrence of specific oligomeric ellagitannins matched well with the phylogeny, suggesting that many of these oligomers can be used as chemotaxonomic signals at least within the plant species found in Finland.
We used a very comprehensive approach to screen the Finnish plant kingdom for the 100 most common species to find the hydrolysable tannin producing plants. This was done by collecting and drying >50 grams of leaf tissue of each species and fractionating 10 grams of the foliar extracts by Sephadex LH-20 gel chromatography into several tannin-free and tannin-rich fractions. After HPLC-DAD and HPLD-ESI-MS analyses of each of the fractions, we were able to reveal how the production of different types of ellagitannin oligomers and monomers in the species followed the known phylogeny of the species (Fig. 1).
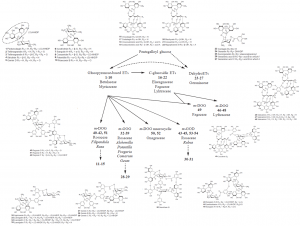
Figure 2. Different types of ellagitannin monomers and oligomers show variable structures and some of them can be specifically linked to certain plant families or even genera. For more details, see Moilanen et al. (2015).
The tannin composition of plants cannot be revealed without hard work
The above types of experiments are very laborious, since tannin identification as such cannot be achieved by using HPLC-DAD and HPLD-ESI-MS only. For that reason we needed to isolate many of the tannins and to identify them by UV, MS and NMR. All this work had been started in our group already in 1998, allowing us to create really powerful tannin library of known structures. These types of high-quality libraries allow us nowadays to find these same tannins in the plant kingdom by using only 20 mg of plant powder in our analyses. That is a huge difference in comparison to the 50 000 mg needed in our first screening experiment (see above). All this is based on the efficient use of accurate and sensitive mass spectrometers attached to UPLC instruments, and on our knowledge of the existing tannin structures in the plant kingdom. More work is needed with these new instruments to verify the above patterns with wider phylogenetic coverage and with wider inclusion of individual hydrolysable tannins.
“I’m losing it – I can see from the leaf shape that this one produces ellagitannins...”
